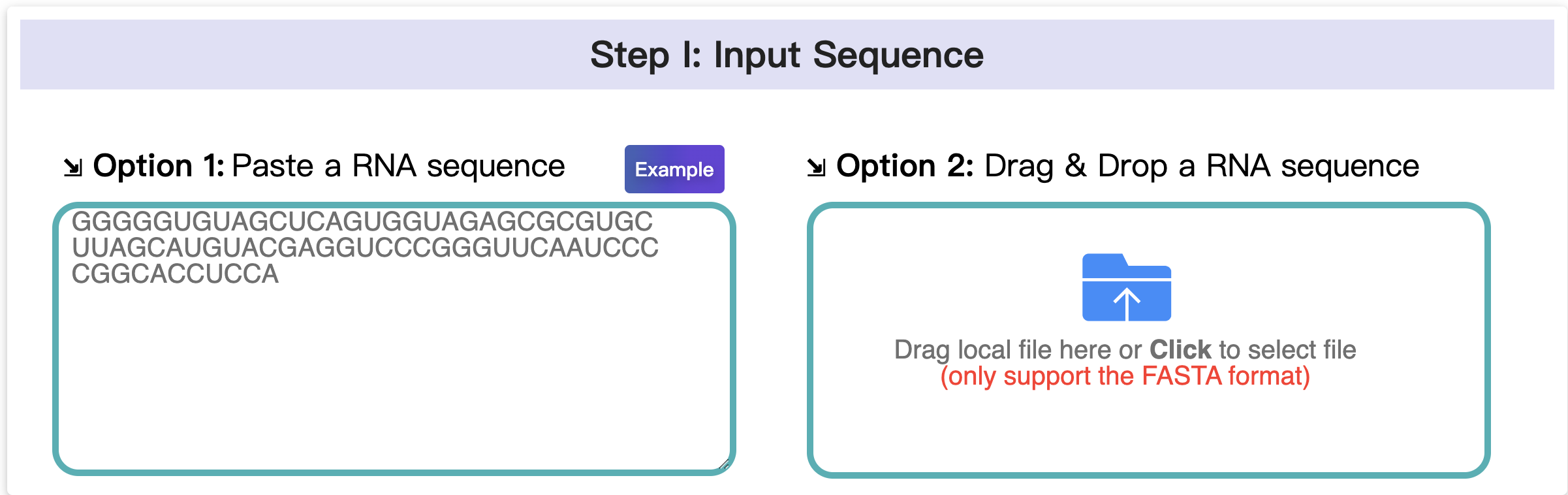
1.1 Input data
In this step, there are two ways to provide your RNA sequence, you can either paste a RNA sequence or upload a FASTA file. Ensure that your RNA sequence contains only the characters A, U, C, and G (no other characters are allowed).
Multiple sequences are not supported. Please provide a single RNA sequence at a time.
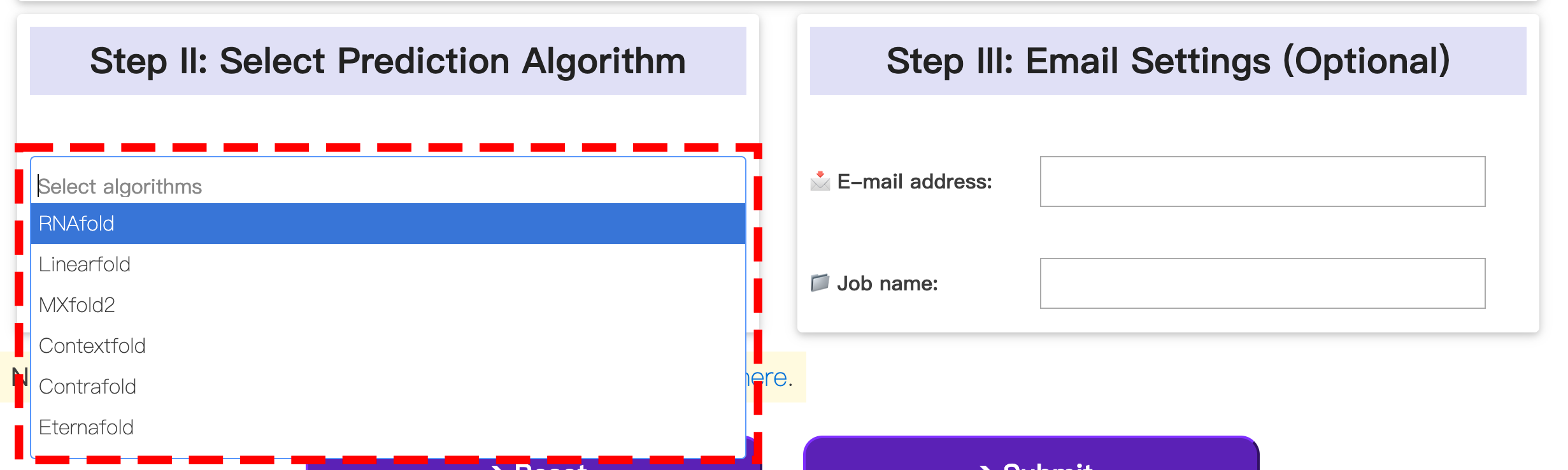
1.2 Settings
In this step, you can select from a variety of algorithms to predict the 2D structure of your RNA sequence. Each algorithm operates on its own distinct principle and may yield different results.
Additionally, users have the option to provide their email address. Once the prediction is complete, they will receive an automatic notification containing the results.
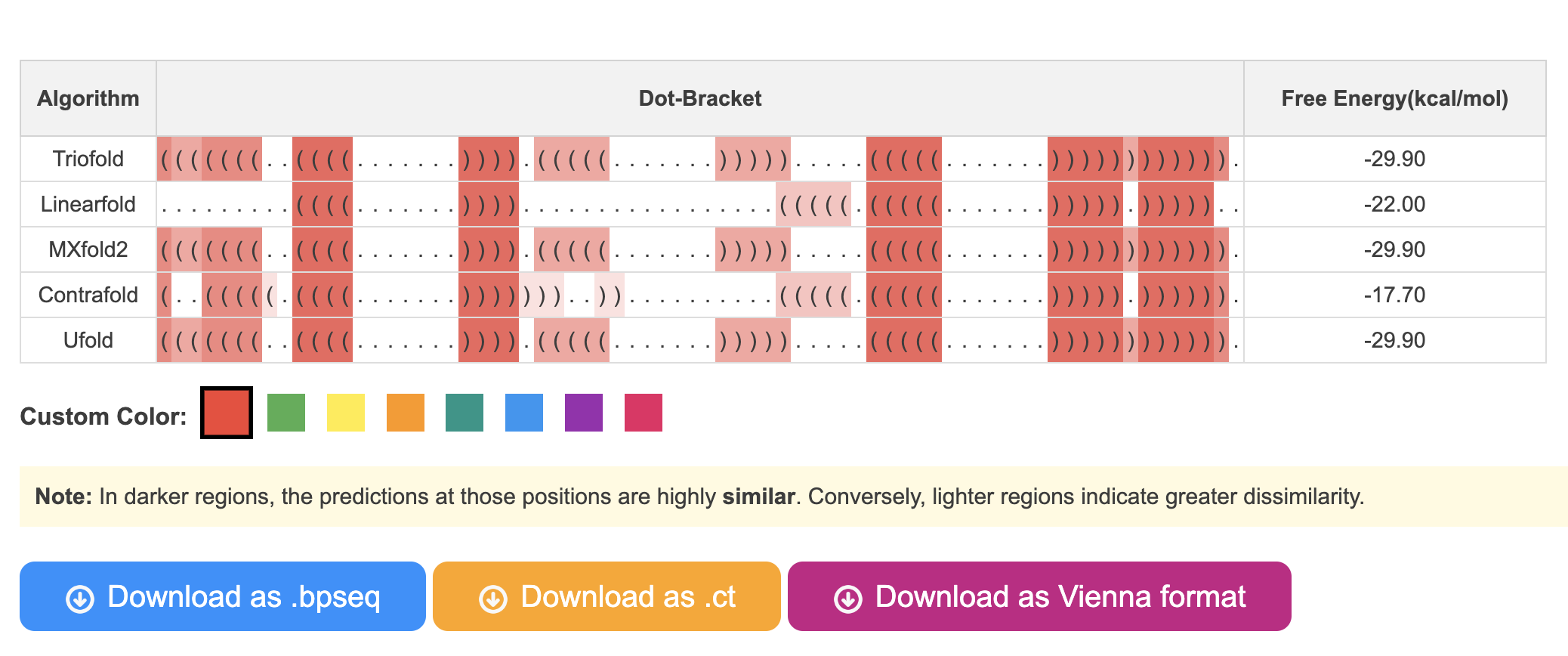
2.1 Dot-bracket outputs
In this section, you are able to see the predicted RNA secondary structures generated by different algorithms. Additionally, the colors representing the frequency of base pair occurrences in various algorithms will vary in shades. You can click on the buttons below to change the color theme.
In addition, results in .ct, .bpseq, and Vienna formats are also provided.
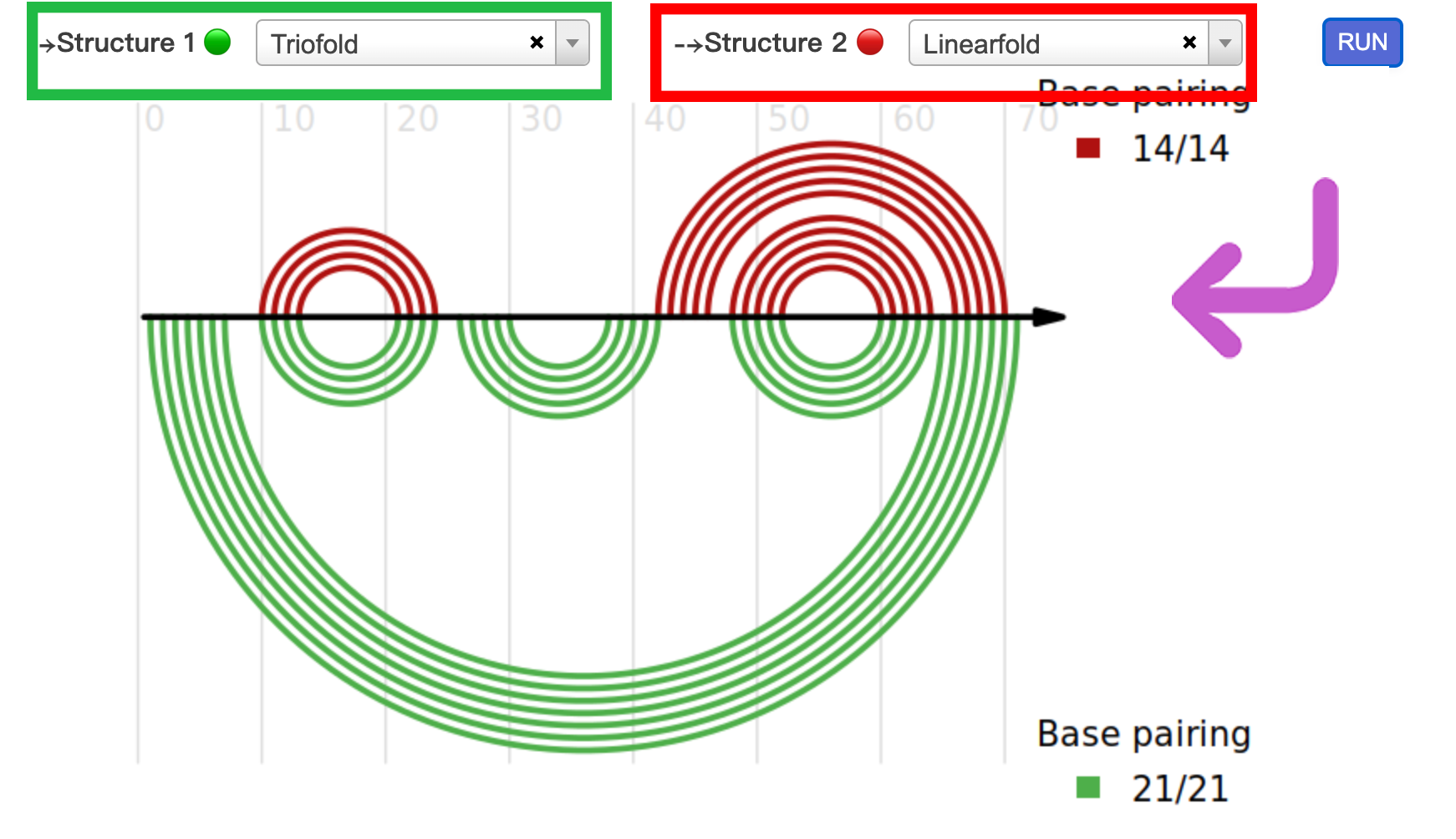
2.2 Pairwise comparision
In this section, we provide users with pairwise comparison of RNA secondary structures. Users can select different algorithms (red and green boxes in the diagram) and click the "run" button to obtain pairwise comparison outputs generated by R-chie.
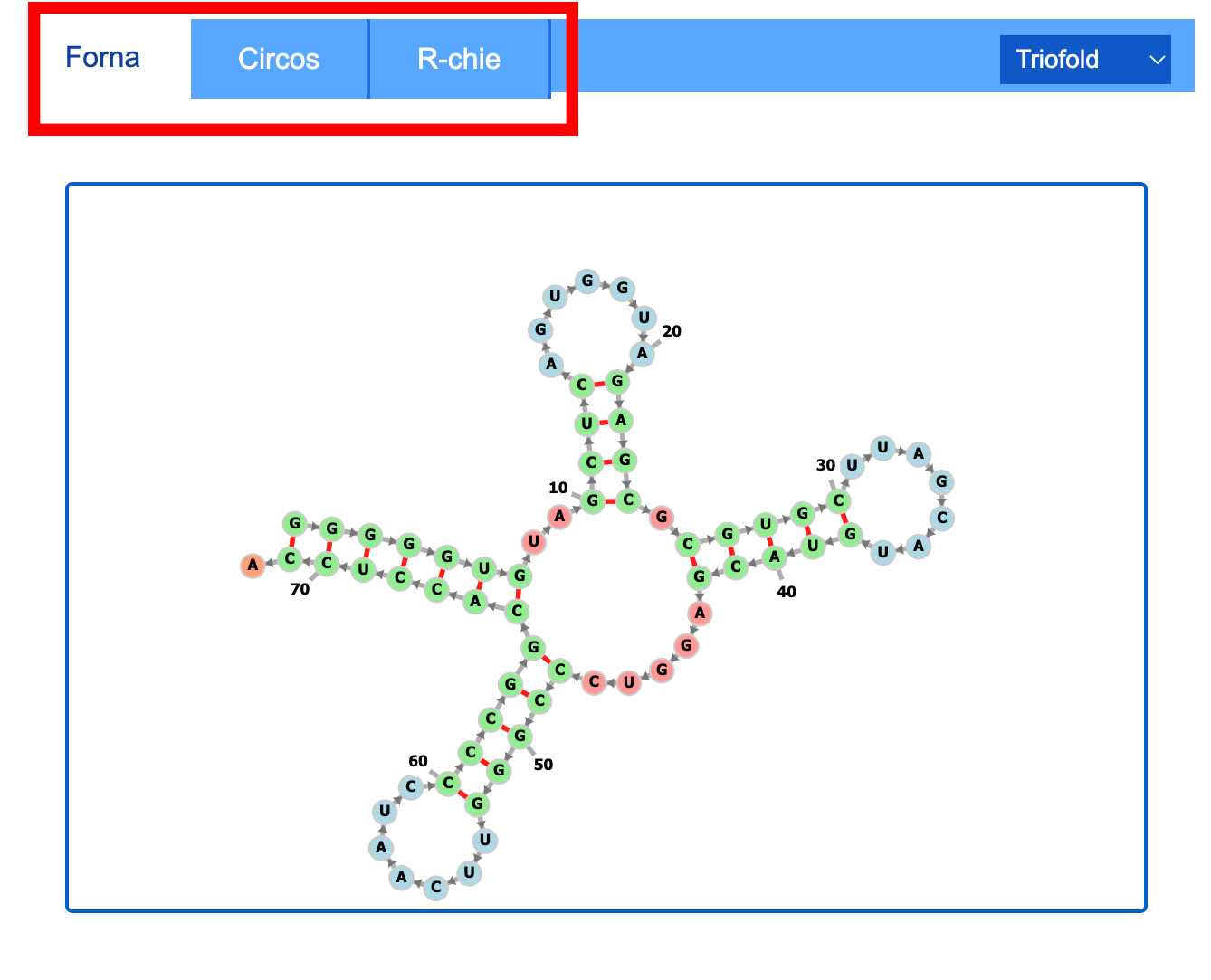
2.3 2D visualization
In this section, you can select from a variety of algorithms to see the 2D structure of your RNA sequence. We offer RNA secondary structure visualization results generated by FORNA, R-chie, and circos.
Additionally, users are provided with a download option for .png formats.
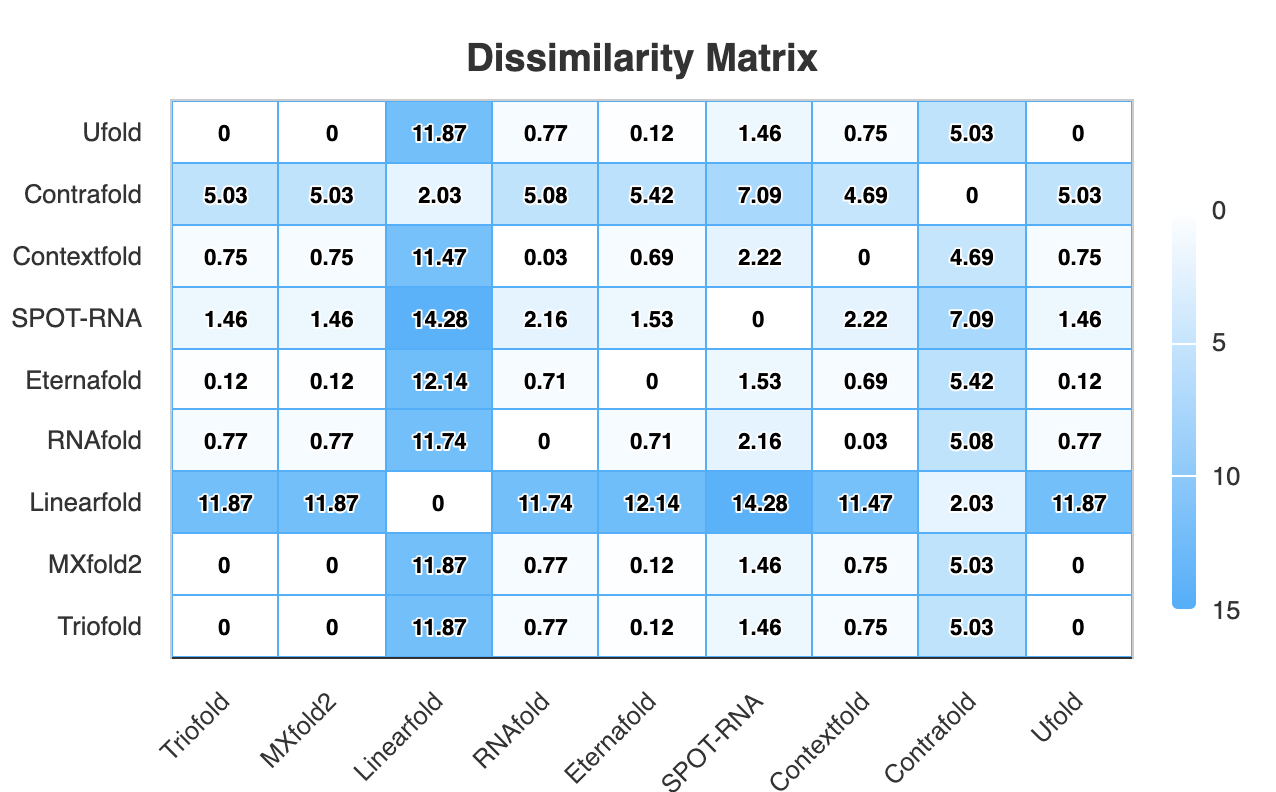
2.4 Dissimilarity analysis
In this section, we utilize Aptamat to characterize the dissimilarity in secondary structure predictions generated by different algorithms. A higher dissimilarity indicates greater disparity between the structures predicted by two algorithms.
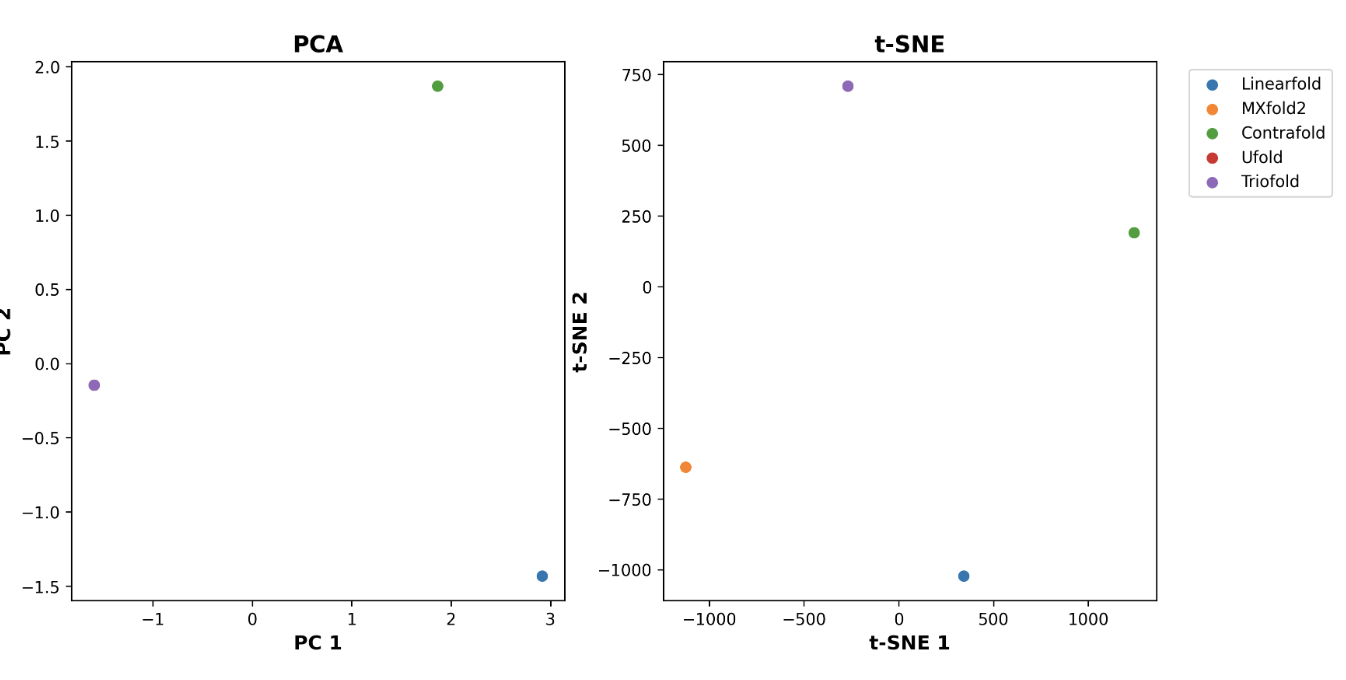
Furthermore, we employ PCA and t-SNE algorithms to cluster the prediction results.
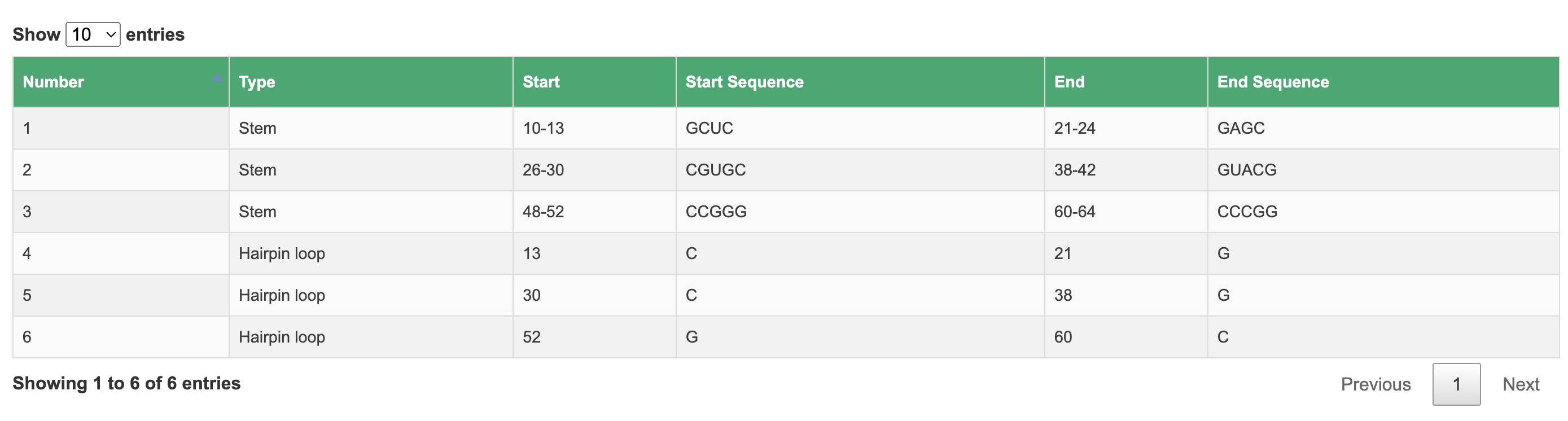
2.5 Motif annotation
In this section, we utilize bpRNA to annotate the structural motifs of RNA.
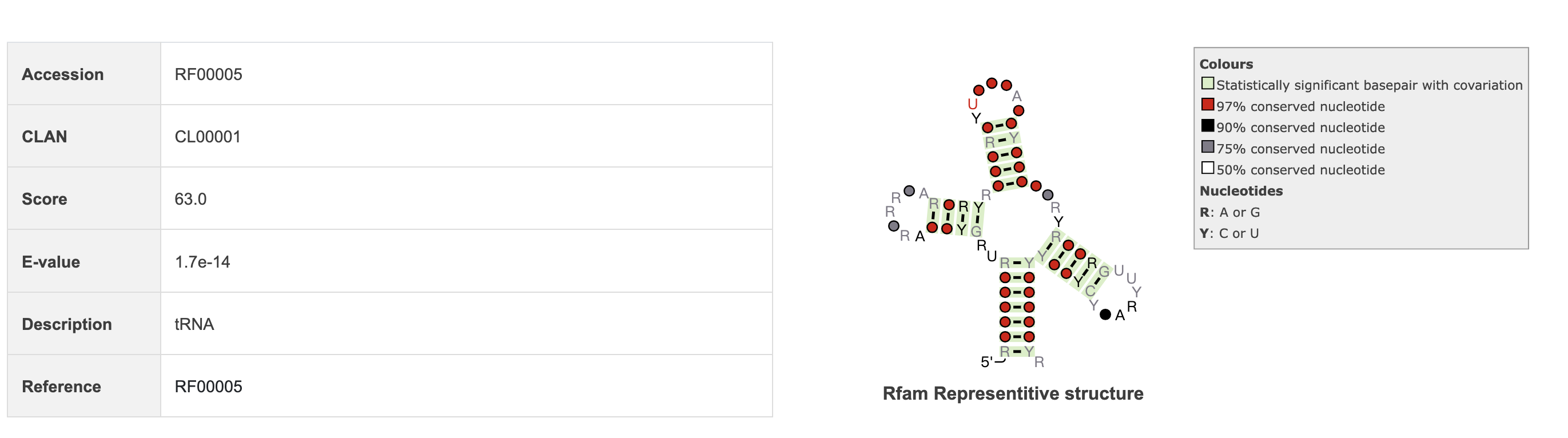
2.6 Rfam annotation
In this section, we utilize Infernal software to annotate RNA sequence and display information such as the sequence's Rfam accession number, description, E-value, and other relevant details.
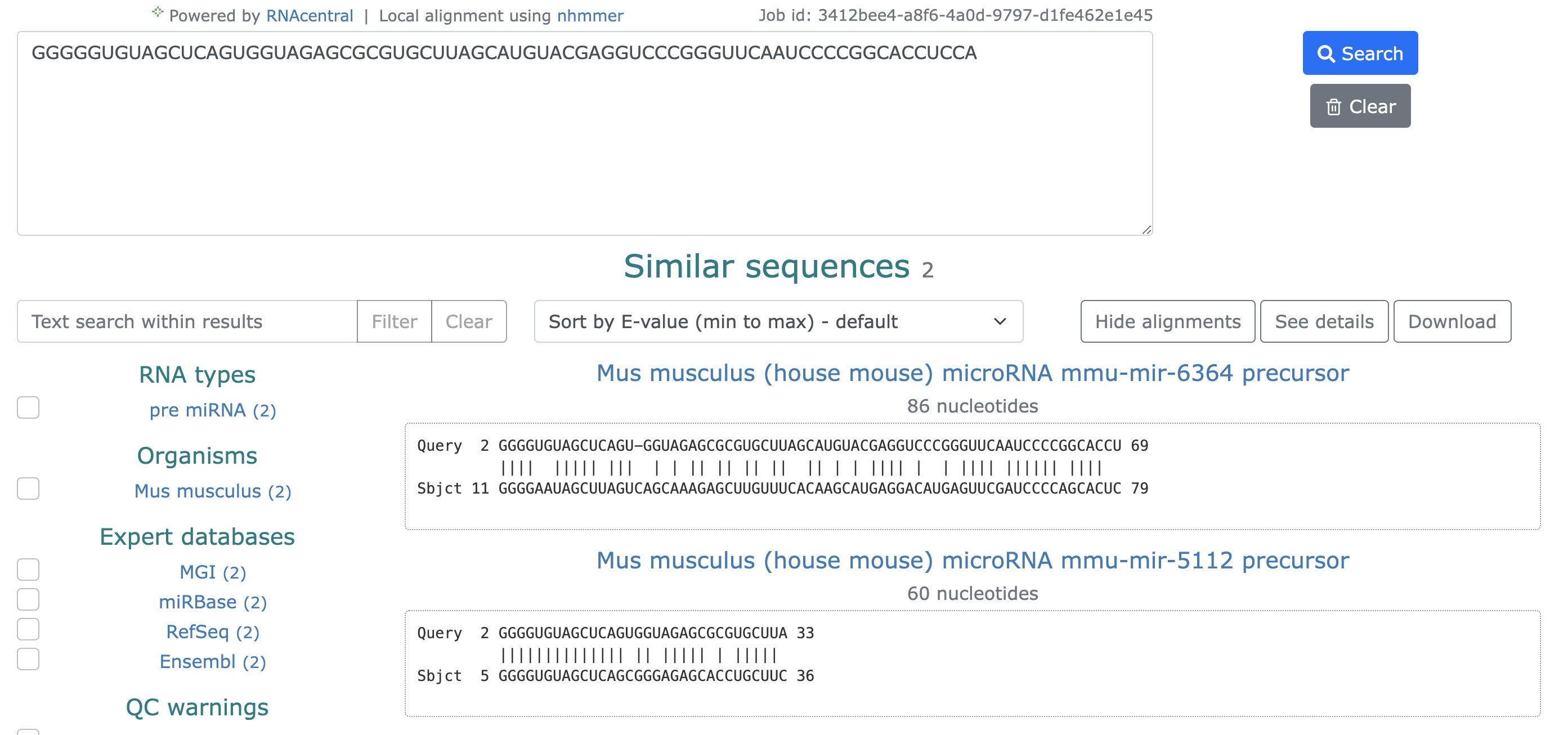
2.7 Similarity search
In this step, we utilize RNAcentral's similarity search to display numerous sequences that are similar to the provided RNA sequence.